Chapter | 2 Web tool
The SysNDD web tool is available from https://sysndd.dbmr.unibe.ch/ on a server hosted at the Department for BioMedical Research (DBMR) (University of Bern) and the web address https://sysndd.org/ redirects to this server.
The web tool uses the Vue.js (v2.6) JavaScript framework with BootstrapVue to generate a Bootstrap v4 website frontend.
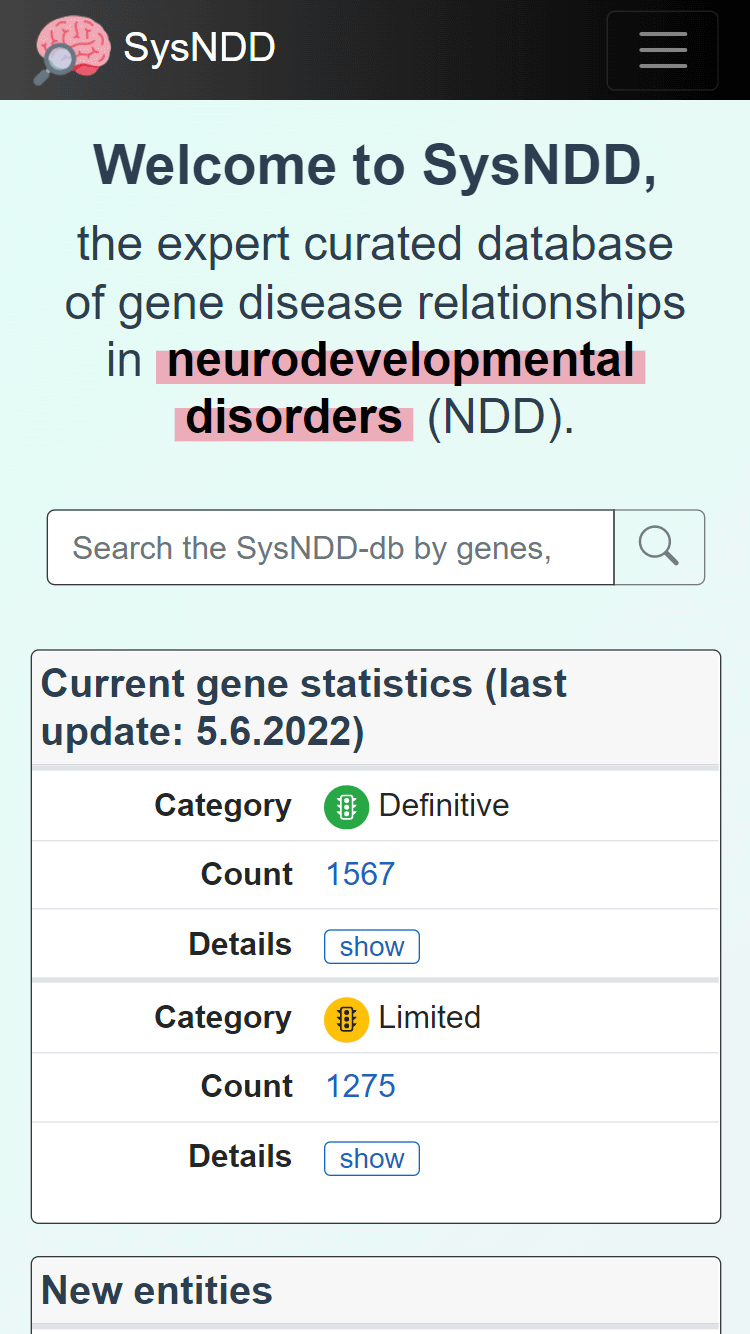
2.1 Landing page
The landing page is designed as simple Bootstrap v4 website with:
- a navigation menu at the top,
- the main site content, which changes with navigation to other routes, and
- a footer navigation bar at the bottom
Screenshot of the landing page with elements marked:
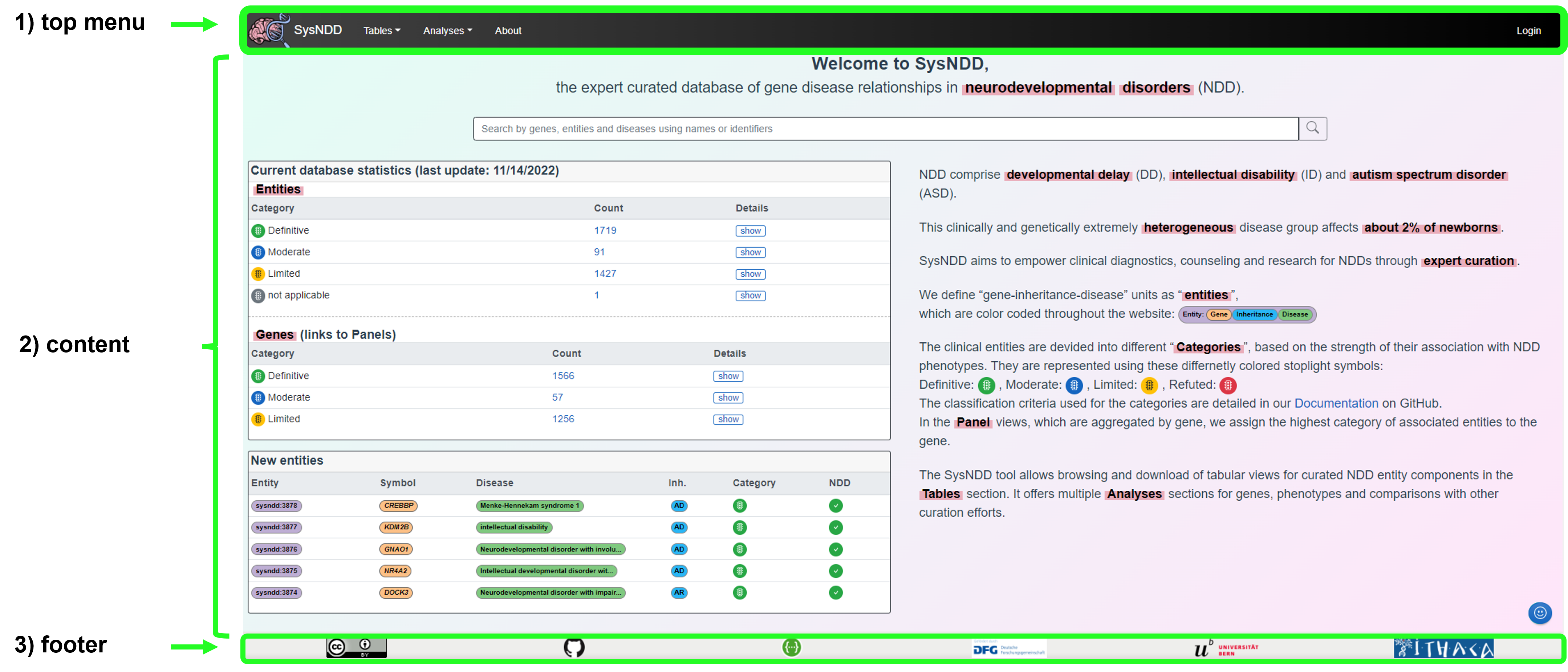
The landing page content includes the following elements to provide a quick overview and allow for easy navigation:
- a centered search input at the top,
- a box (left side top) with current gene statistics divided by association category and inheritance patterns (Details),
- a box (left side bottom) showing a table of the five last entities entered into the database,
- an explanatory text on the right.
2.2 Main navigation menu
The main navigation allows quick access to all sub-pages.
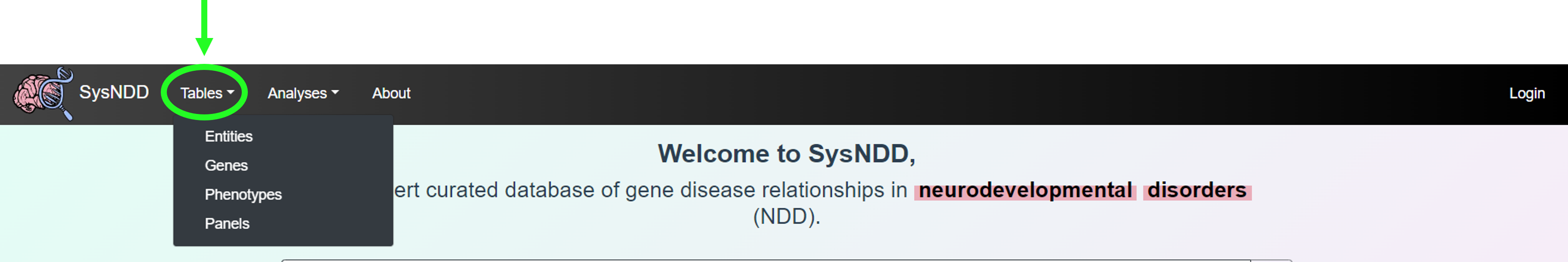
The Tables button triggers a dropdown menu with links to:
- Entities table view
- Genes table view
- Phenotypes table view
- Panels table view
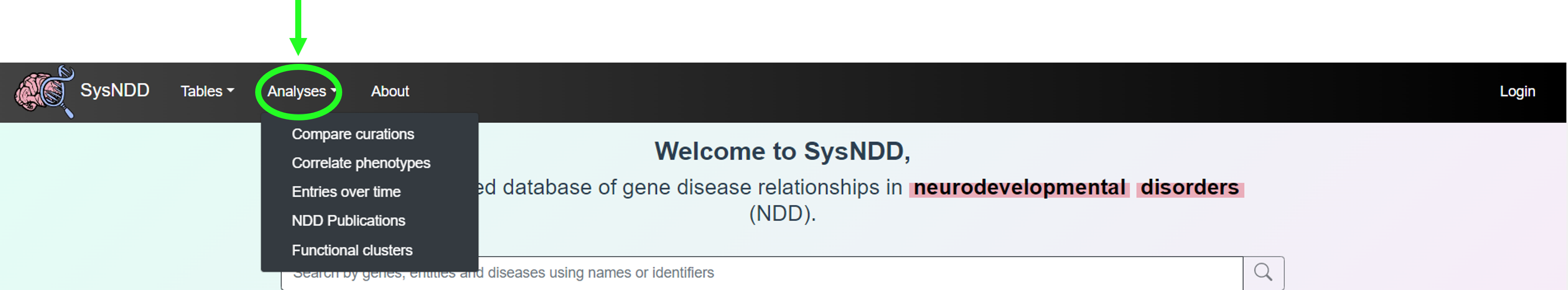
The Analyses button triggers a dropdown menu with links to:
- Compare curations view
- Correlate phenotypes view
- Entries over time view
- NDD Publications view
- Functional clusters view
The About button directs you to further information on:
- SysNDD and its creators
- Citation Policy
- Support and Funding
- News and Updates
- Credits and acknowledgement
- Help and FAQ
- Disclaimer
- Contact

If not on the landing page, a search bar also appears on the navigation menu.
If not logged in, the right side of the menu shows a button which directs you to the Login page. When logged in as a registered user the menu shows your username and additional links to page views depending on your user rights:
2.3 Footer navigation menu
The footer navigation shows logos with links to:
- the license applied to SysNDD
- our GitHub repository
- the SysNDD API view
- the DFG funder website
- the website of the University of Bern hosting our server
- the ERN-ITHACA website

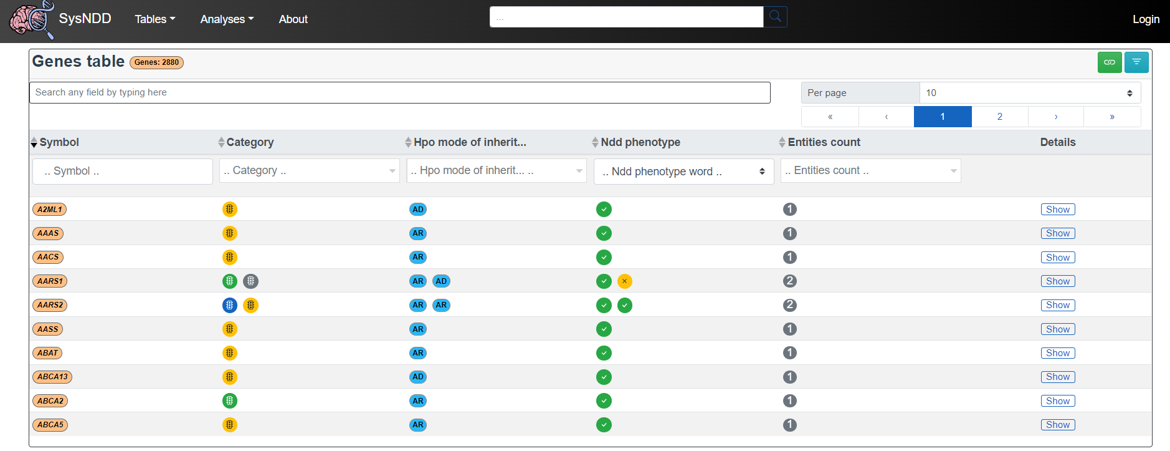
2.4 Table views
We provide tabular representations with search, filtering, sorting and pagination functionality for different aspects of the entity concept.
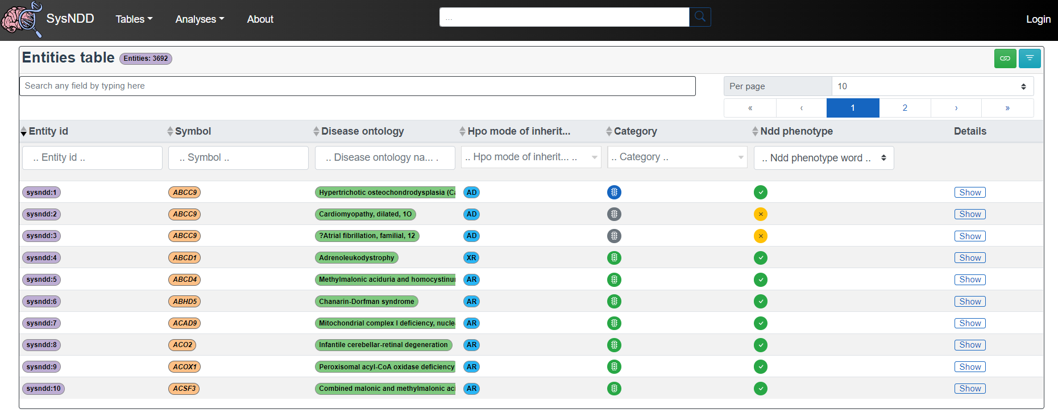
2.4.1 Entities table
The Entities table is intended to provide an overview centered on the entity concept.
2.4.3 Phenotypes table
The Phenotypes table provides the possibility to filter for phenotype combinations annotated to the entities.
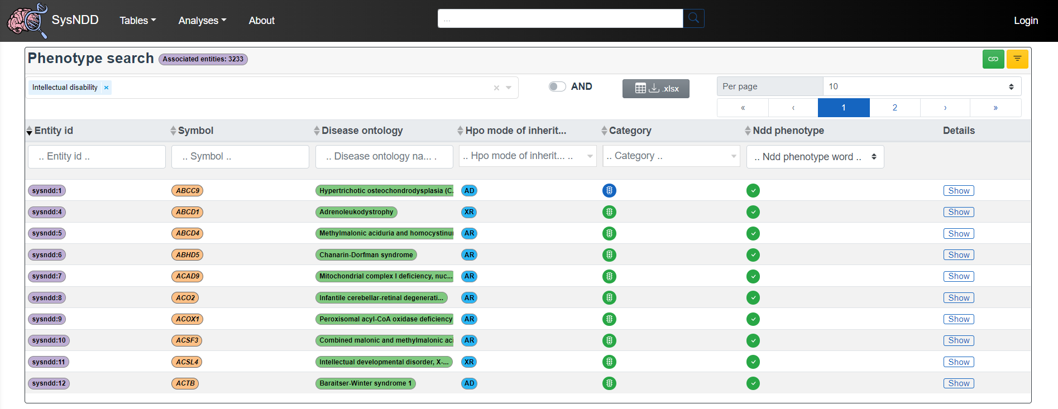
The ‘AND/ OR’ switch allows the user to change the logic how phenotype combinations are requested:
- AND: only entities having all selected phenotypes annotated are shown
- OR: all entities having any of the selected phenotypes annotated are shown
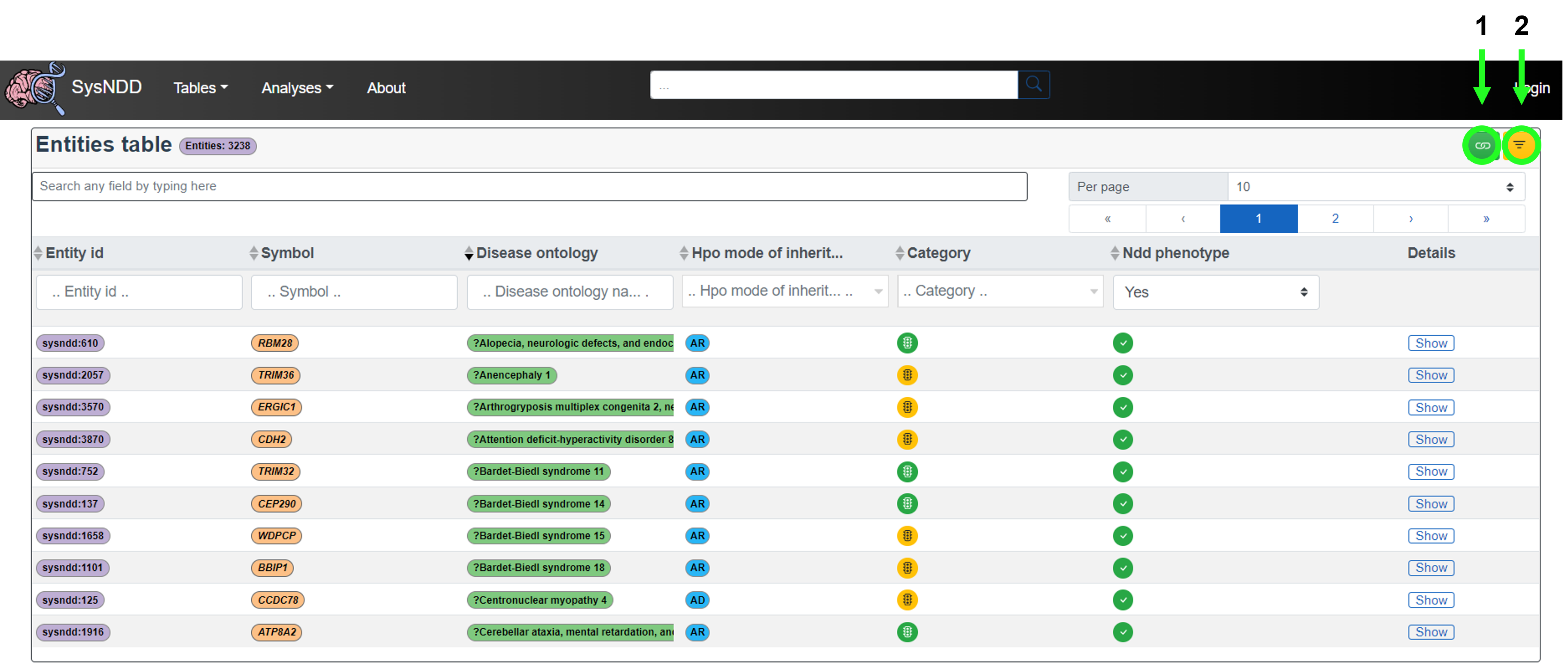
The Entities, Genes and Phenotypes tables all have the additional two features (top right corner):
- Green icon that allows the user to copy the link to the page
- Yellow icon that allows the use to remove all filters on the table (icon turns blue when all filters are off)
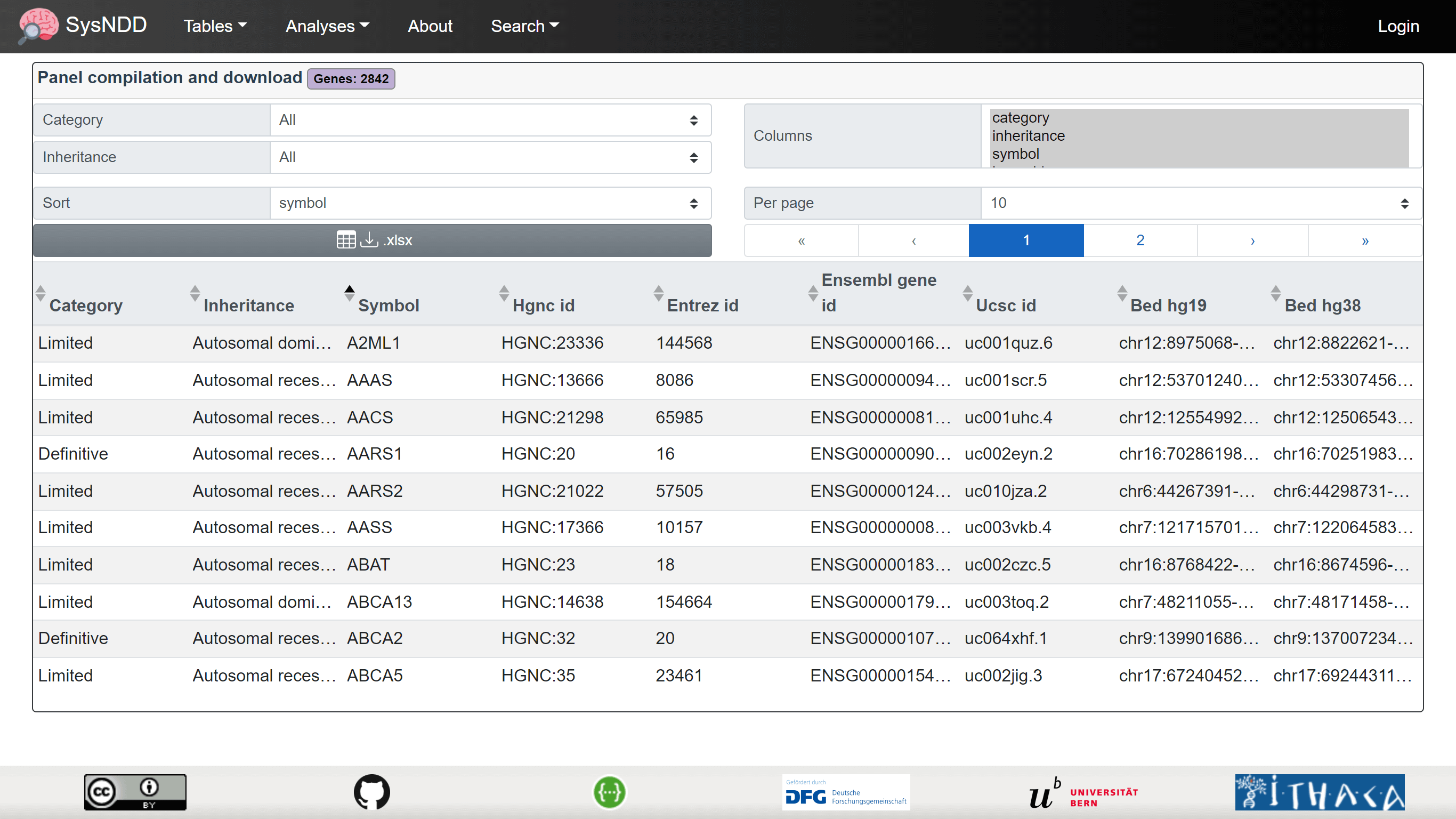
2.4.4 Panels table
The Panels table is intended for users to be able to create lists of NDD-associated genes. Additionally, the columns in the lists can be configured. Finally, the configuration can be downloaded as an Excel file with information on the exact query in the meta sheet and the requested information in the data sheet. These files can then be used as ‘virtual panels’ to filter genetic variants derived from high-throughput sequencing in external analysis tools.
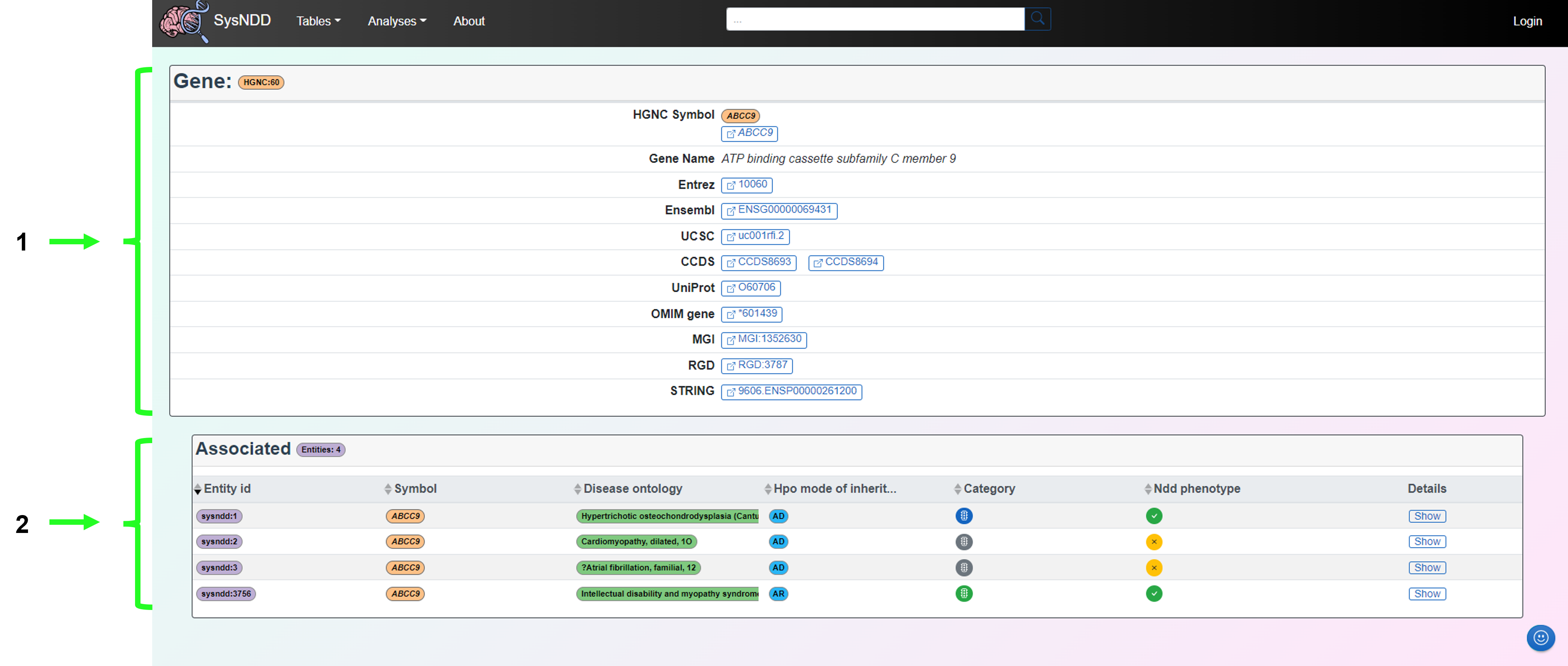
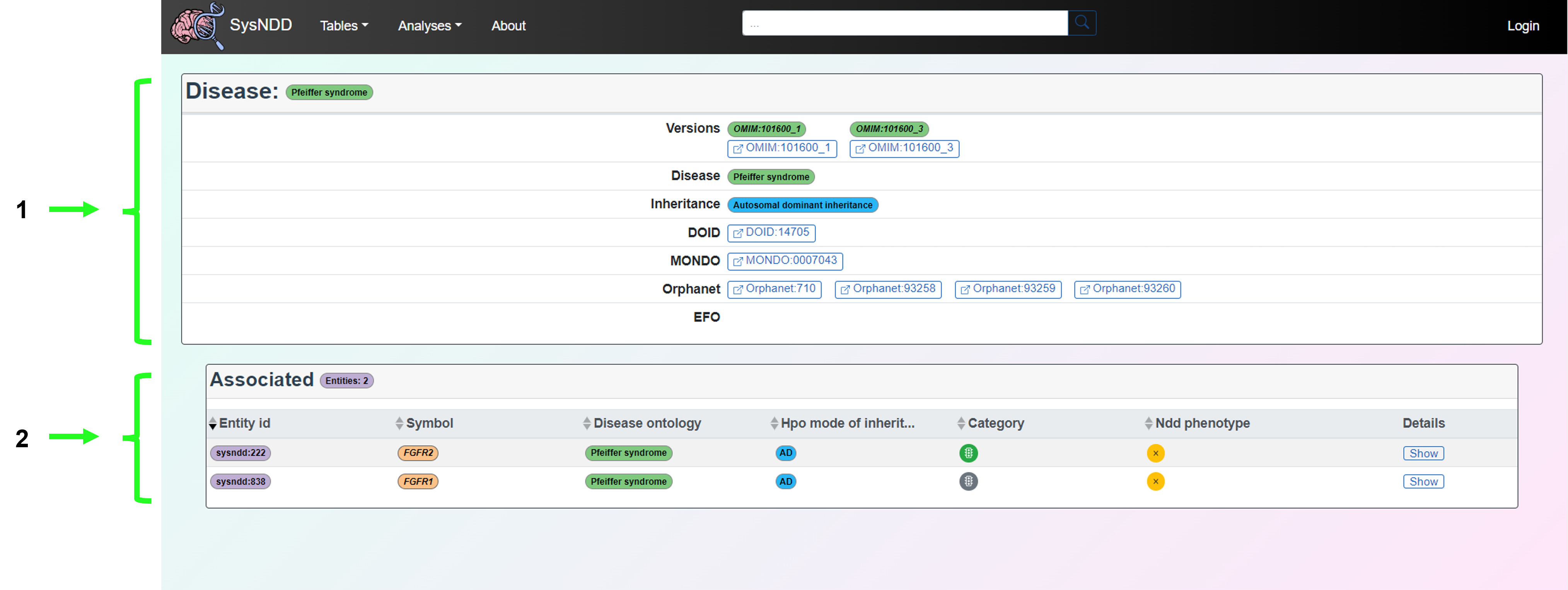
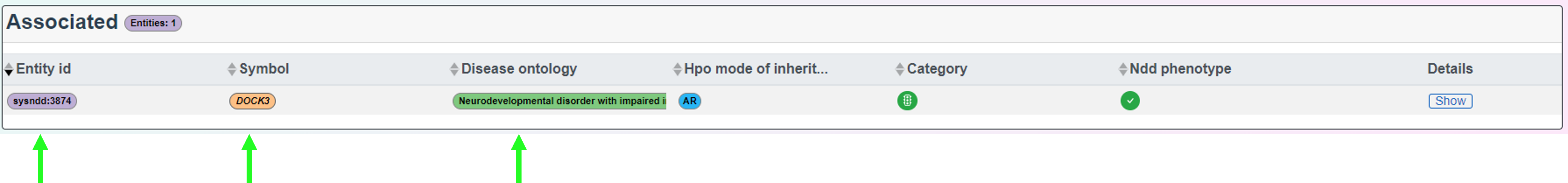
2.5 Single entry pages
Single entry pages refer to the directed pages associated with each entity, gene or disease ontology. These are accessed by clicking on the entity (purple), gene (orange) or disease (green) buttons associated with each entry.
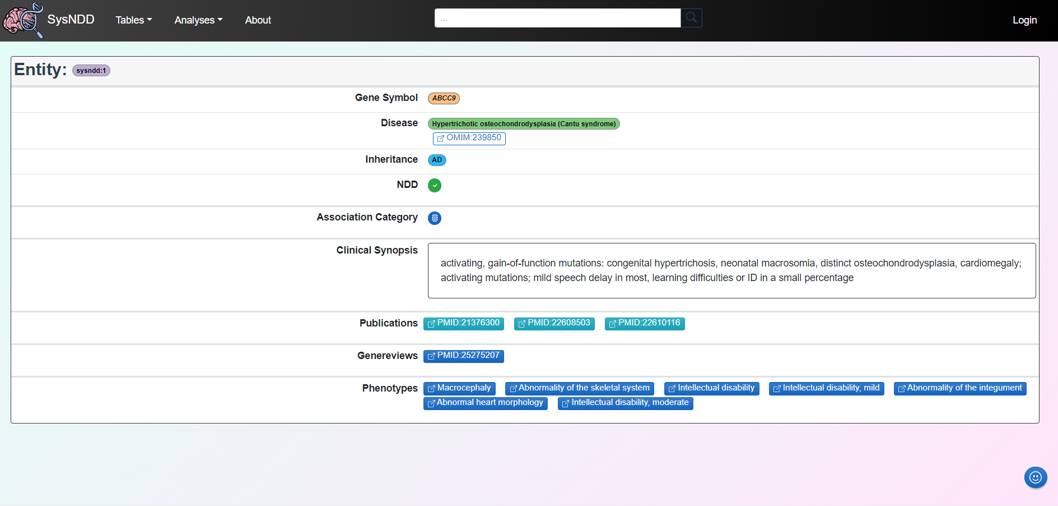
2.5.1 Entity
The Entity page aims to provide information on the gene, disease (with the OMIM link), inheritance pattern and NDD status associated with a particular entity. The synopsis and phenotypes of the disease as well as the relevant links to publications (including GeneReviews if available) are also provided.
2.6 Analyses views
The Analysis views are intended to provide the user with a more comprehensive picture of SysNDD comparisons with other curations, phenotype correlations, SysNDD entries over time, NDD publications, and functional gene clusters.
2.6.1 Compare curations
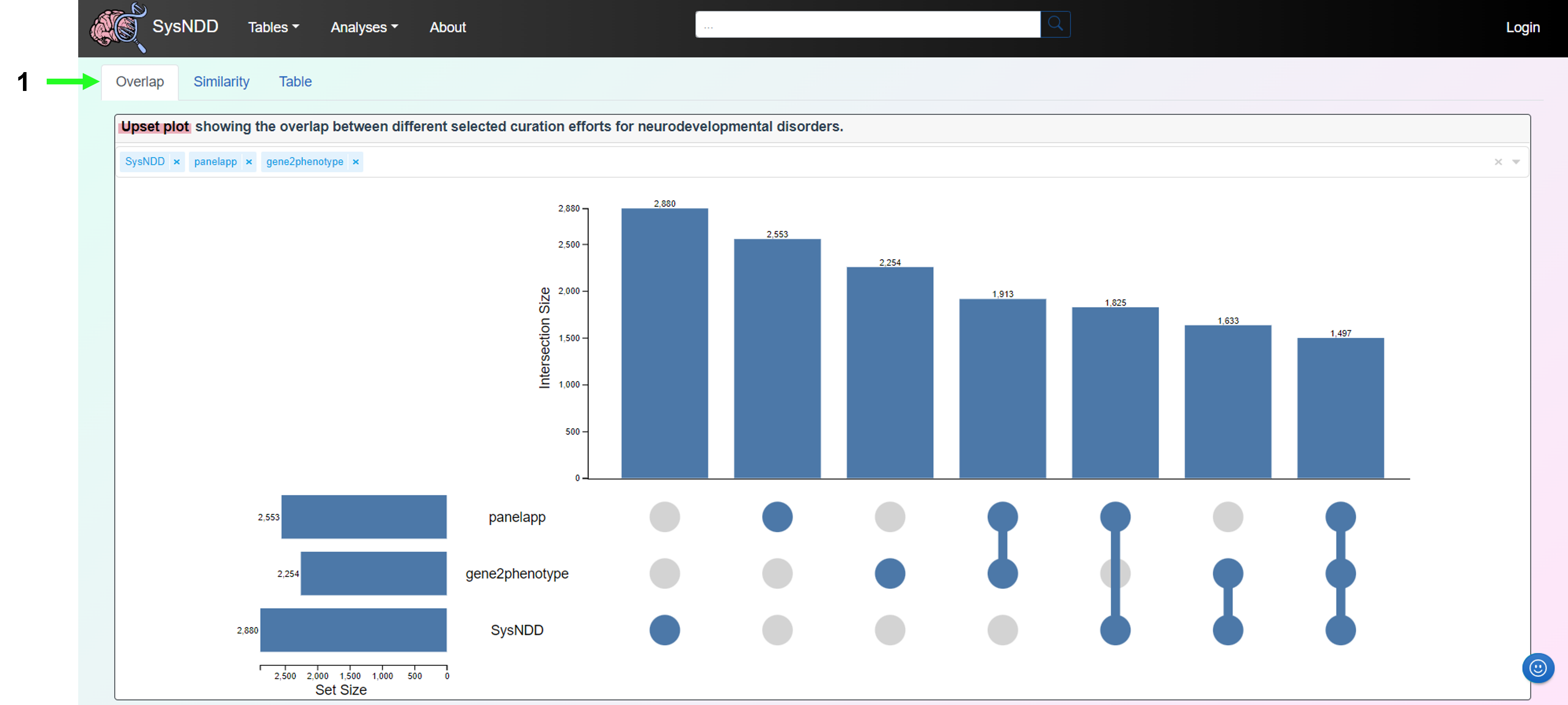
The Compare curations view is composed of three tabs:
- Overlap
- Similarity
- Table
The Overlap tab includes an upset plot (alternative for venn diagrams, please compare: upset.app) to show the overlap between SysNDD and other selected NDD curation efforts.
The Similarity tab includes a matrix plot of the cosine similarity between different curation efforts for neurodevelopmental disorders.
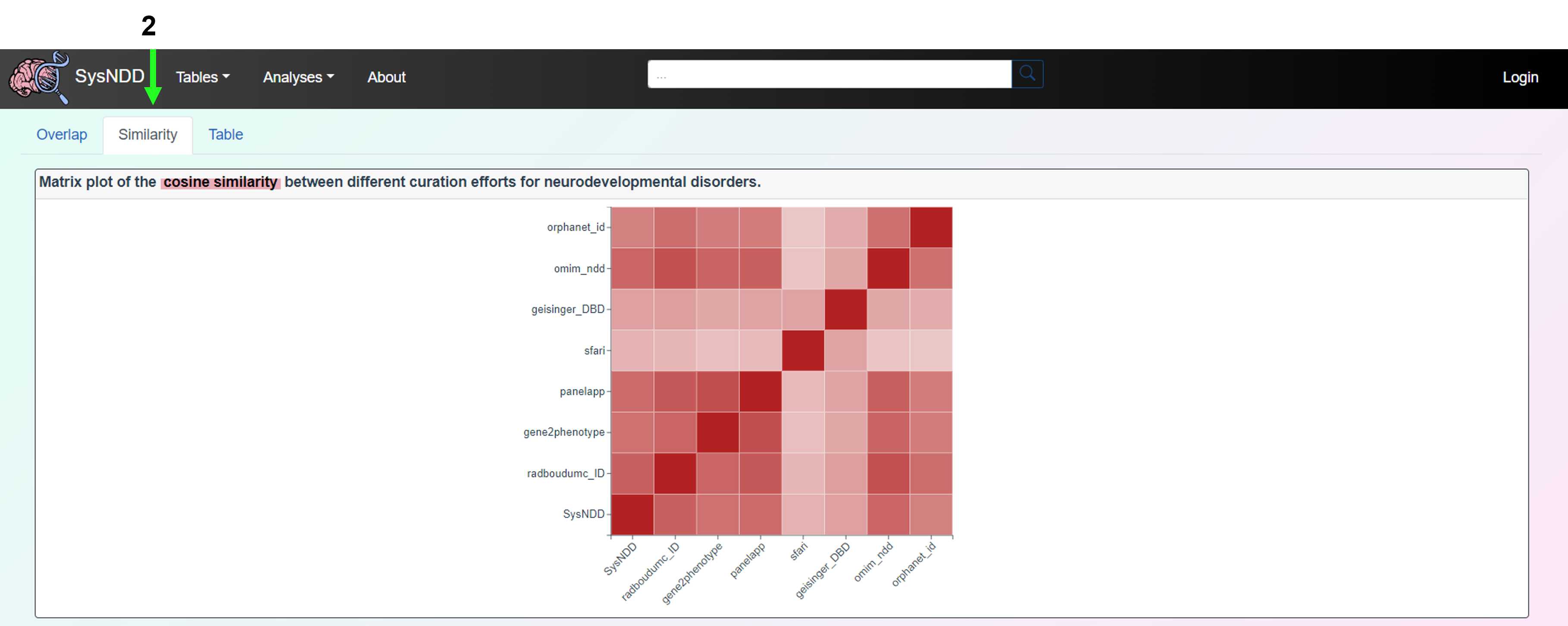
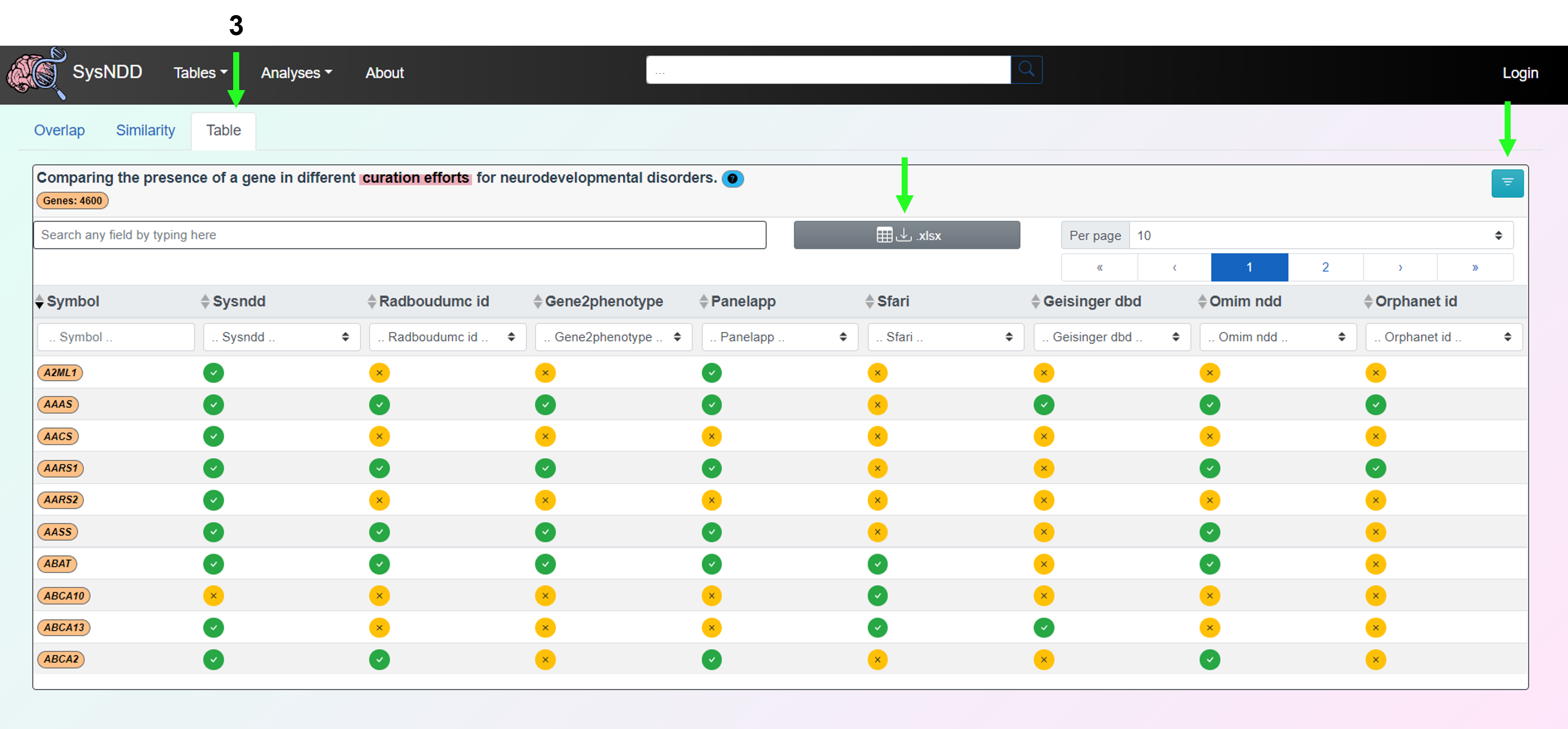
The Table tab presents a table format for comparing a gene’s presence across various neurodevelopmental disorder curation initiatives. The comparison table can also be downloaded as an Excel file using this tab. In the upper right corner, there is a button to remove the filter.
2.6.2 Correlate phenotypes
Correlate phenotypes view is composed of three tabs:
- Phenotype correlogram
- Phenotype counts
- Phenotype clustering
The Phenotype correlogram tab displays a matrix of correlations of different phenotypes.

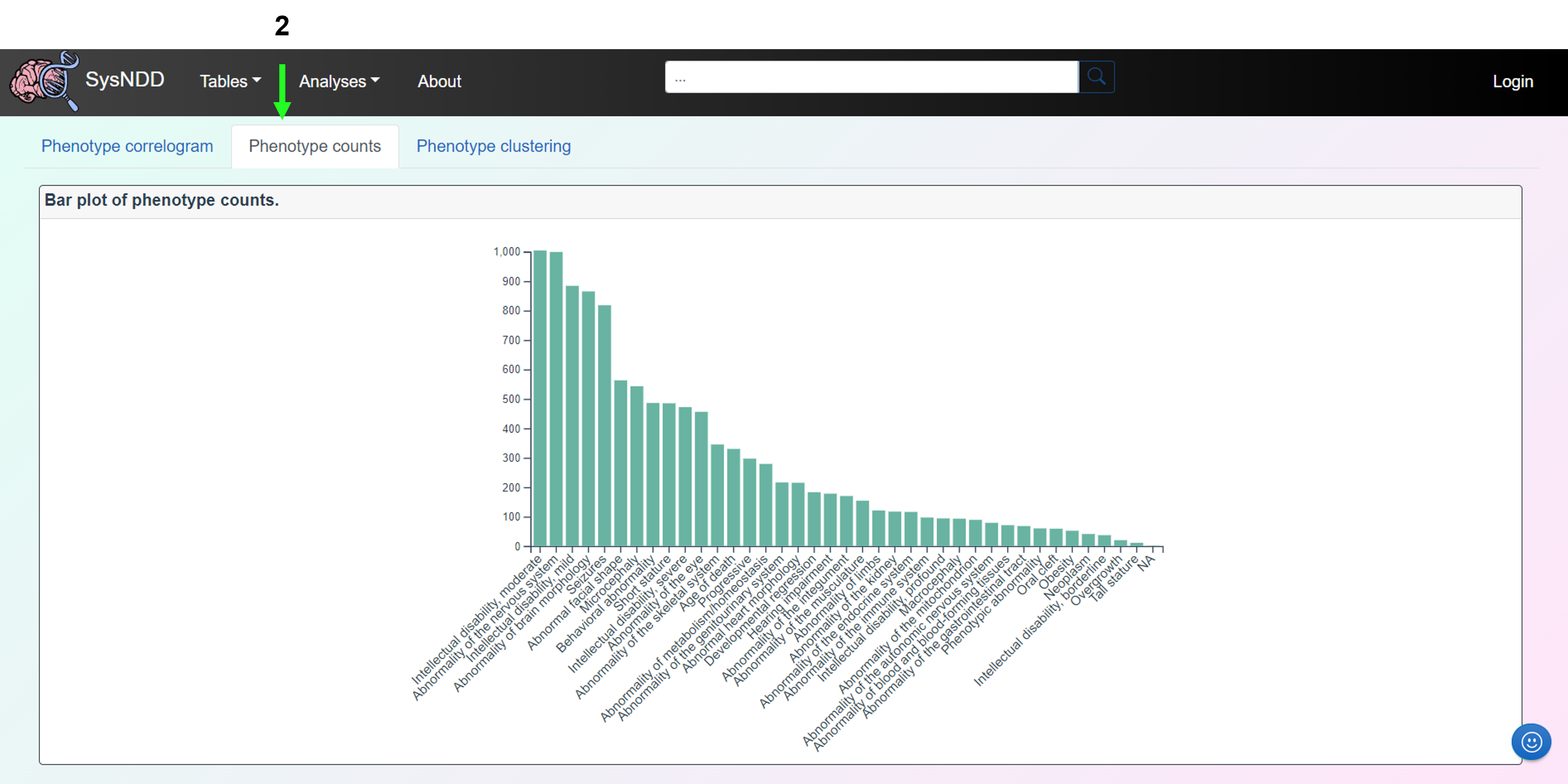
The Phenotype counts tab shows a bar plot of phenotype counts.
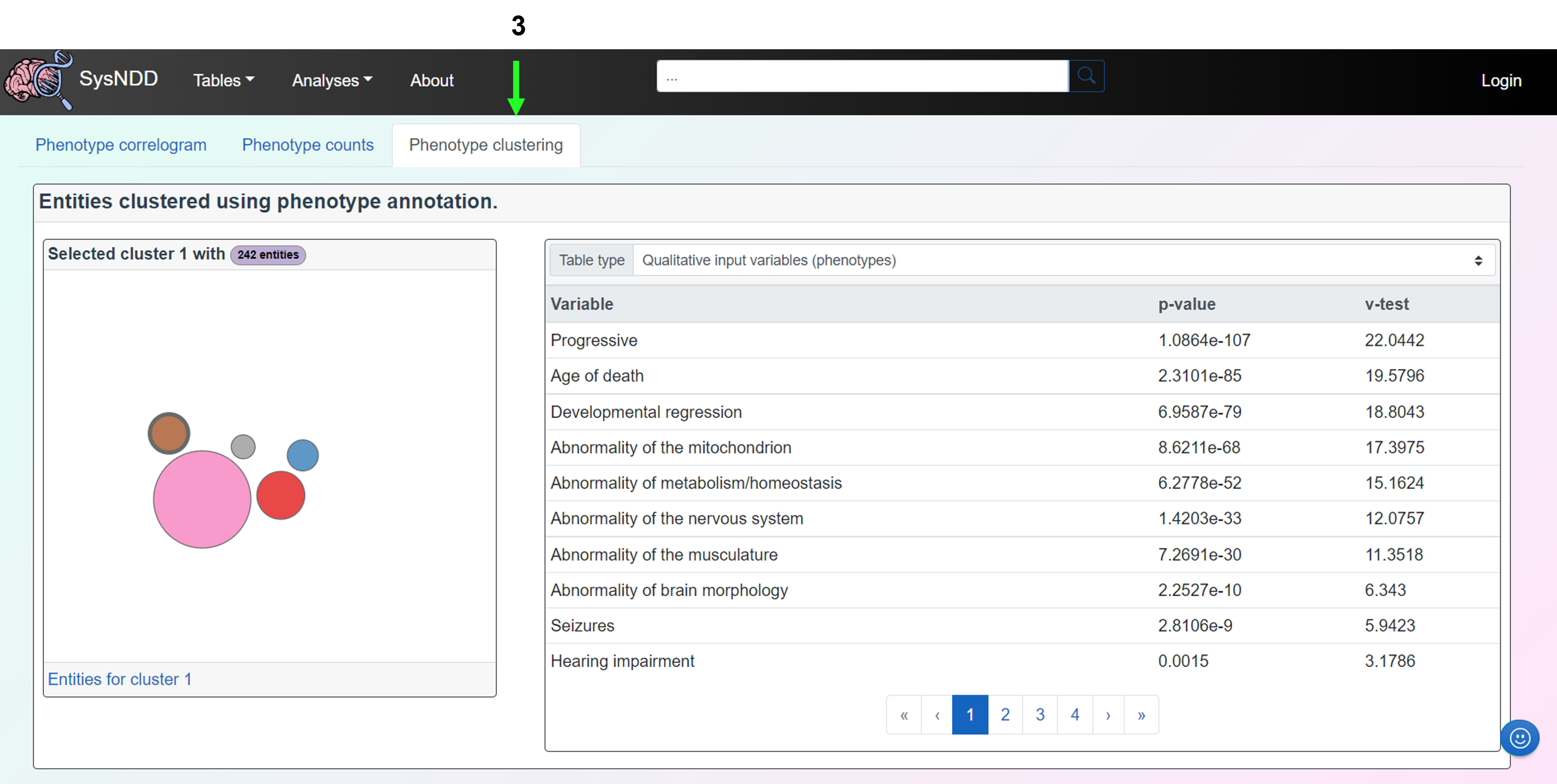
The Phenotype clustering tab shows clusters of entities based on the manually curated phenotype annotations from SysNDD. Multiple correspondence analysis (MCA) is performed utilizing the “MCA” function from the FactoMineR R package, with the number of dimensions retained set to 15, the qualitative supplementary variables set to inheritance terms, and the quantitative supplementary variables set to phenotype counts divided into ID-related and non-ID-related phenotypes (indicator of “syndromicity”). Then, hierarchical clustering is performed using the “HCPC” function from the FactoMineR package. By clicking on the different colored bubbles on the panel to the left, the user can select the respective clusters. When clicking a cluster the entity count is displayed in the upper part along the cluster name. The link in this panel’s lower section leads to a view of the Entity table that is restricted to the entities in the selected cluster. The right-hand panel displays a table containing either (1) the Qualitative input variables representing the phenotypes, (2) the Qualitative supplementary variables (independent) representing inheritance patterns, or (3) the Quantitative supplementary variables (independent) representing the count of phenotypes, all with variable name, p-values, and v-test values.
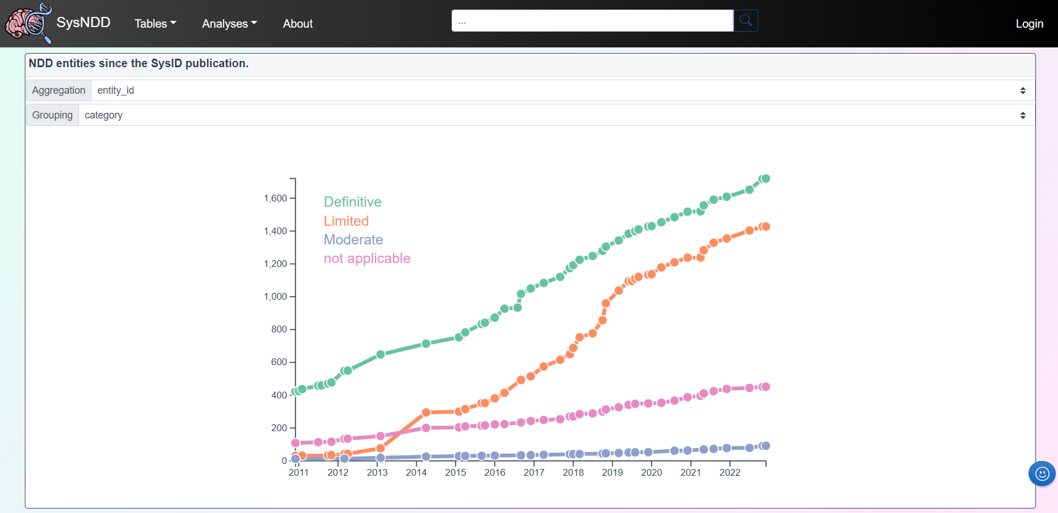
2.6.3 Entries over time
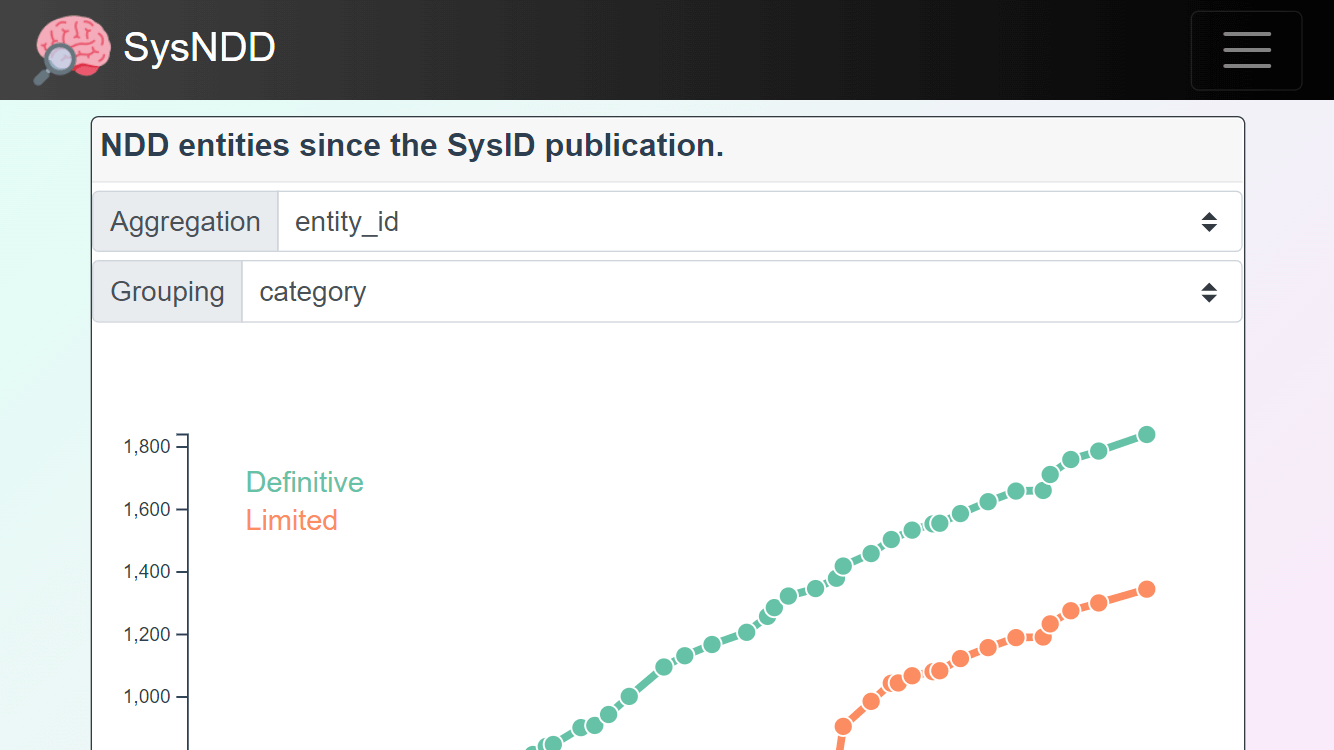
The Entries over time view displays the changes in NDD entity numbers since curation began. The plot can be aggregated by either genes or entities and categorized according to inheritance or category.
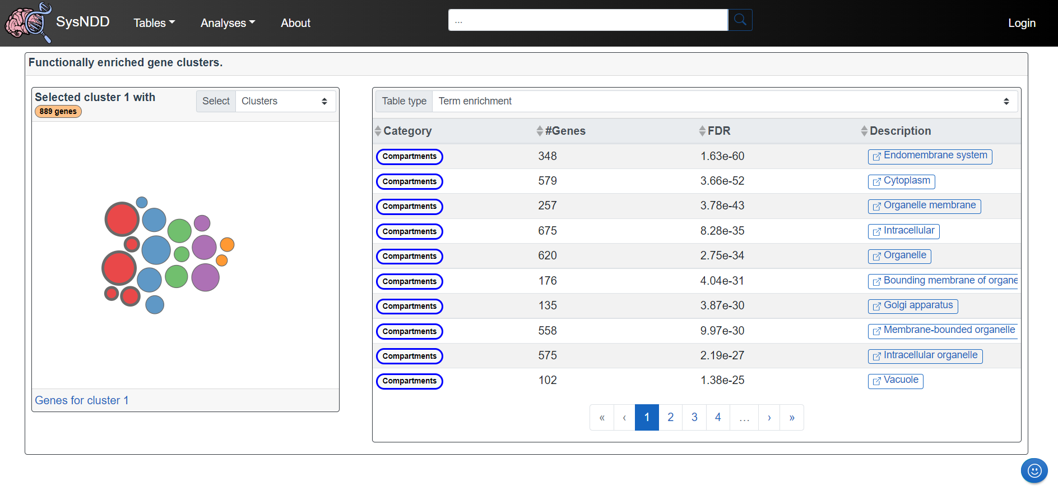
2.6.5 Functional clusters
The Functional clusters view displays gene clusters of functionally enriched interacting proteins, along with their corresponding ontology annotations. Using the “get_clusters” function from the STRINGdb R package and the “walktrap” clustering algorithm from the igraph R package, we perform clustering. By clicking on the different colored bubbles on the panel to the left, the user can select the respective main- or sub-clusters. When clicking a cluster the gene count is displayed in the upper part along the cluster name. The link in this panel’s lower section leads to a view of the Gene table that is restricted to the genes in the selected cluster. The right-hand panel displays a table with either (1) the Term enrichment including the ontology annotations, the number of enriched genes, the FDR-corrected p-value, and a link to the corresponding ontology term or (2) the gene Identifiers with links to the respective single entry page and to the STRING website of the protein.
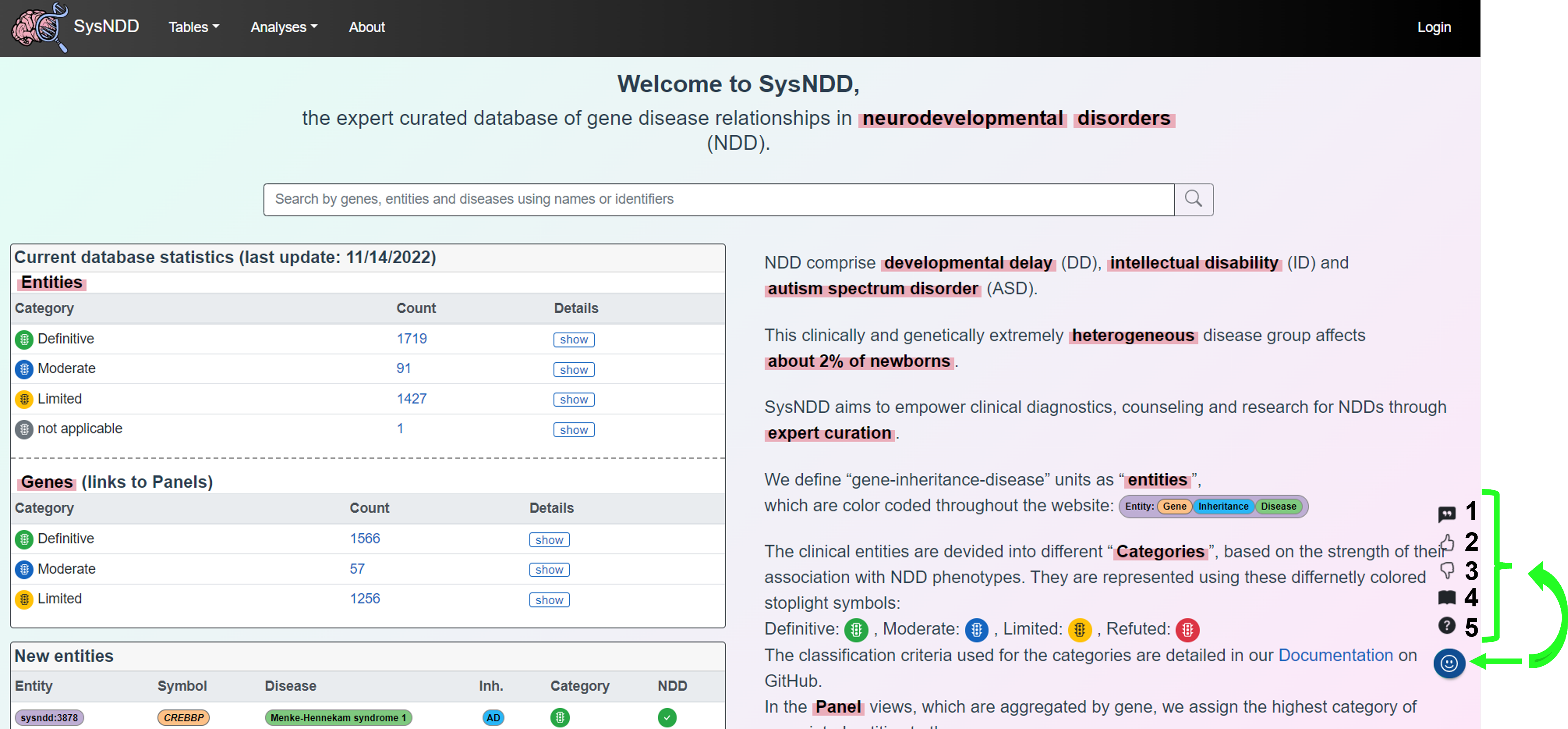
2.7 About page
The website’s About page provides general information about the project, such as its creators, funding sources, the status of updates, and how to get help.
2.8 Help & Feedback
On each page of SysNDD, a smiley face button (in blue, bottom right) points users to the following help and feedback features:
- Button 1 allows the user to cite the page (quotation mark icon), copies the recommended citation text to the clipboard and automatically creates a snapshot of teh website in the internet archive for reproducibility (Wayback Machine - Internet Archive)
- Button 2 directs the user to a form for positive feedback (thumbs up icon)
- Button 3 directs the user to a form for improvement suggestions (thumbs down icon)
- Button 4 directs the user to the SysNDD documentation (book icon)
- Button 5 directs the user to the SysNDD GitHub discussions page for questions and help (question mark icon)
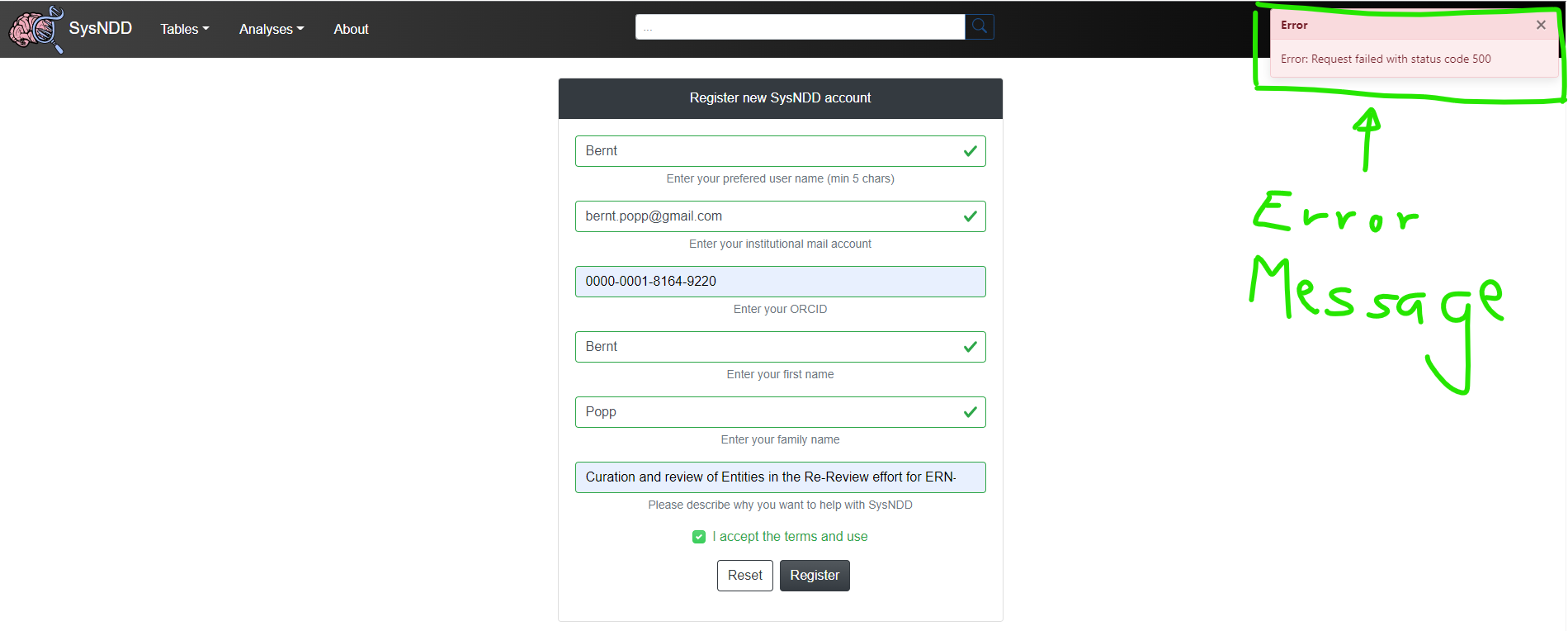
2.9 Reporting bugs, problems and making feature requests
If you have technical problems using SysNDD or requests regarding the data or functionality, please contact us at support [at] sysndd.org.
When reporting bugs and problems please describe in detail on which page you encountered the issue and what input you gave. Provide screenshots including the error message (upper right corner).
You can also save the console logs to aid us in identifying the problem and send the log file in your mail:
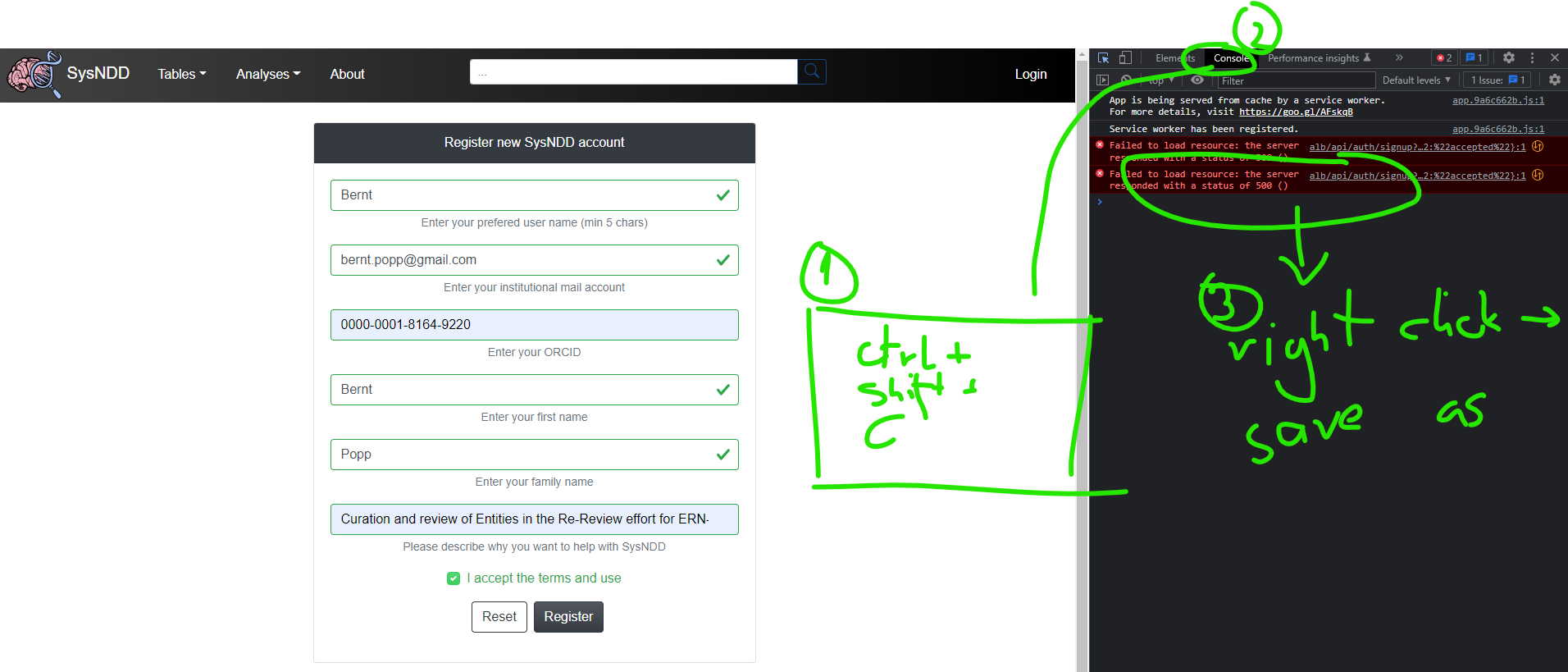
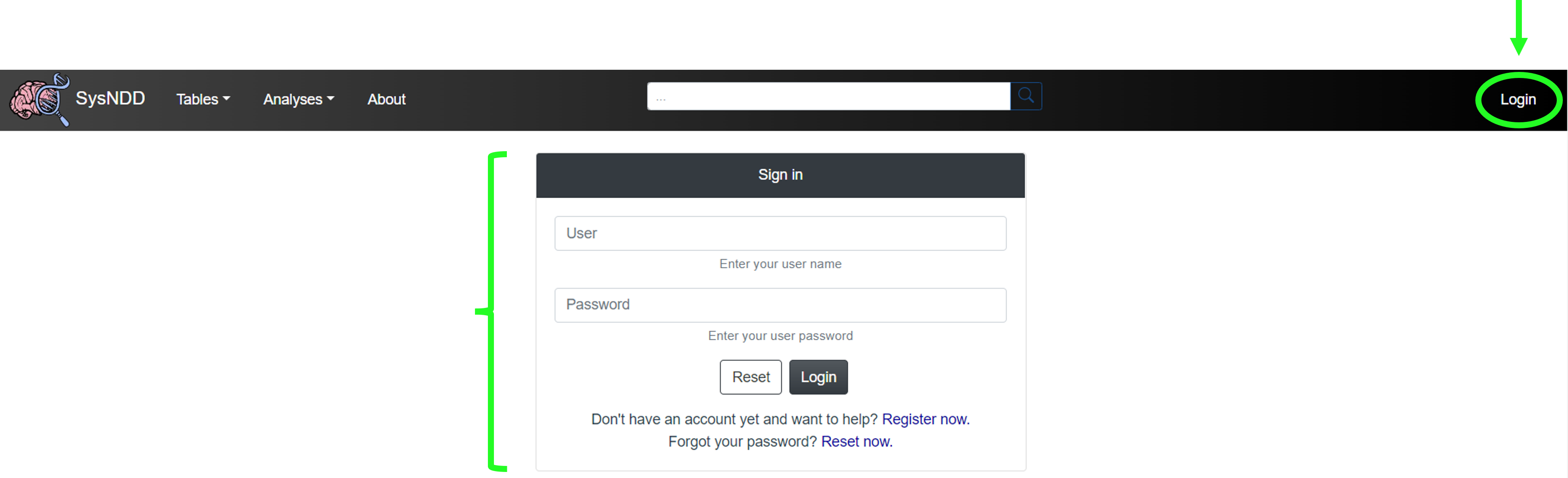
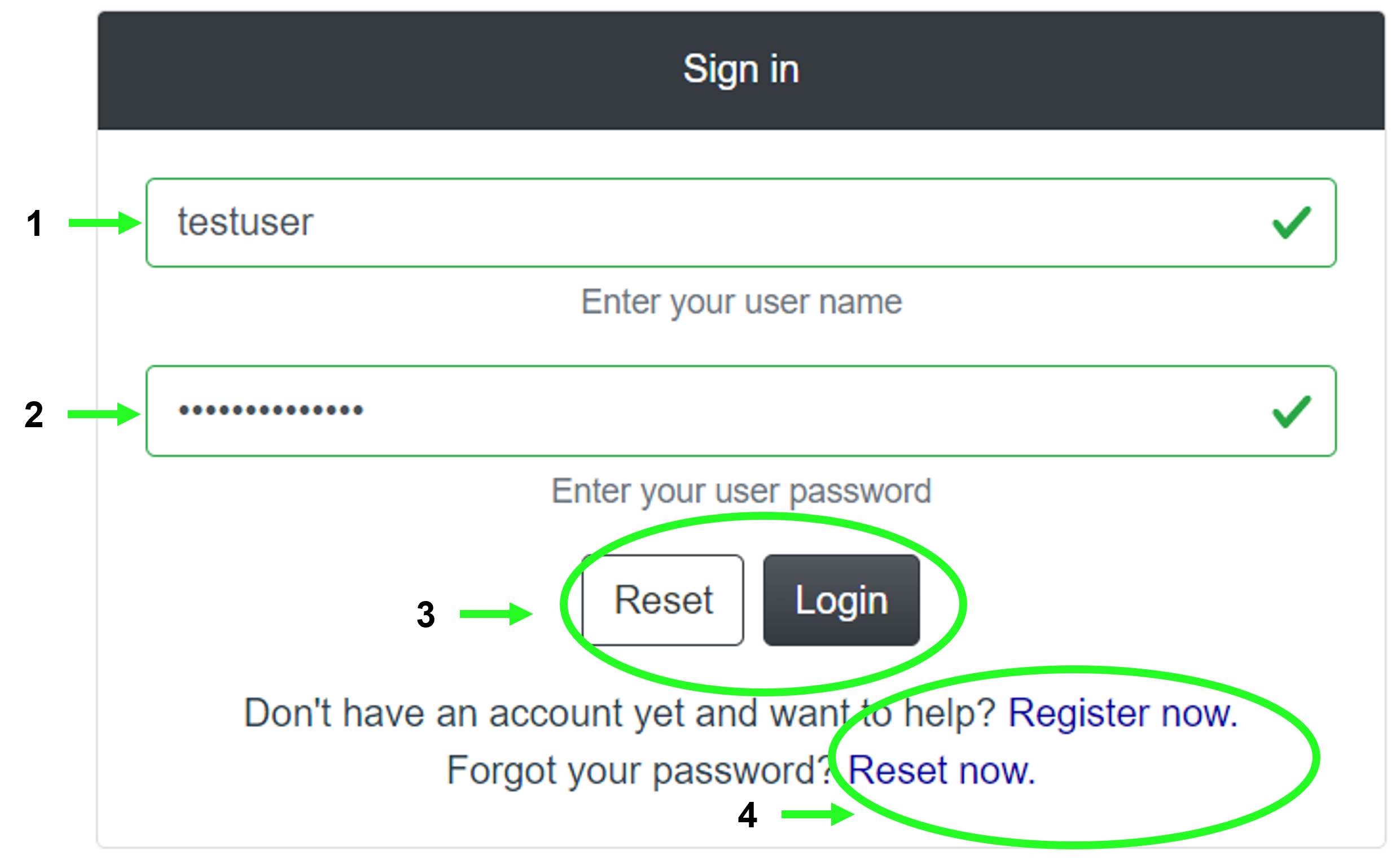
2.10 Login page
The Login page shows a simple form with inputs for the (1) user name and their (2) password, (3) buttons to reset the form and (4) links to registration and password reset.
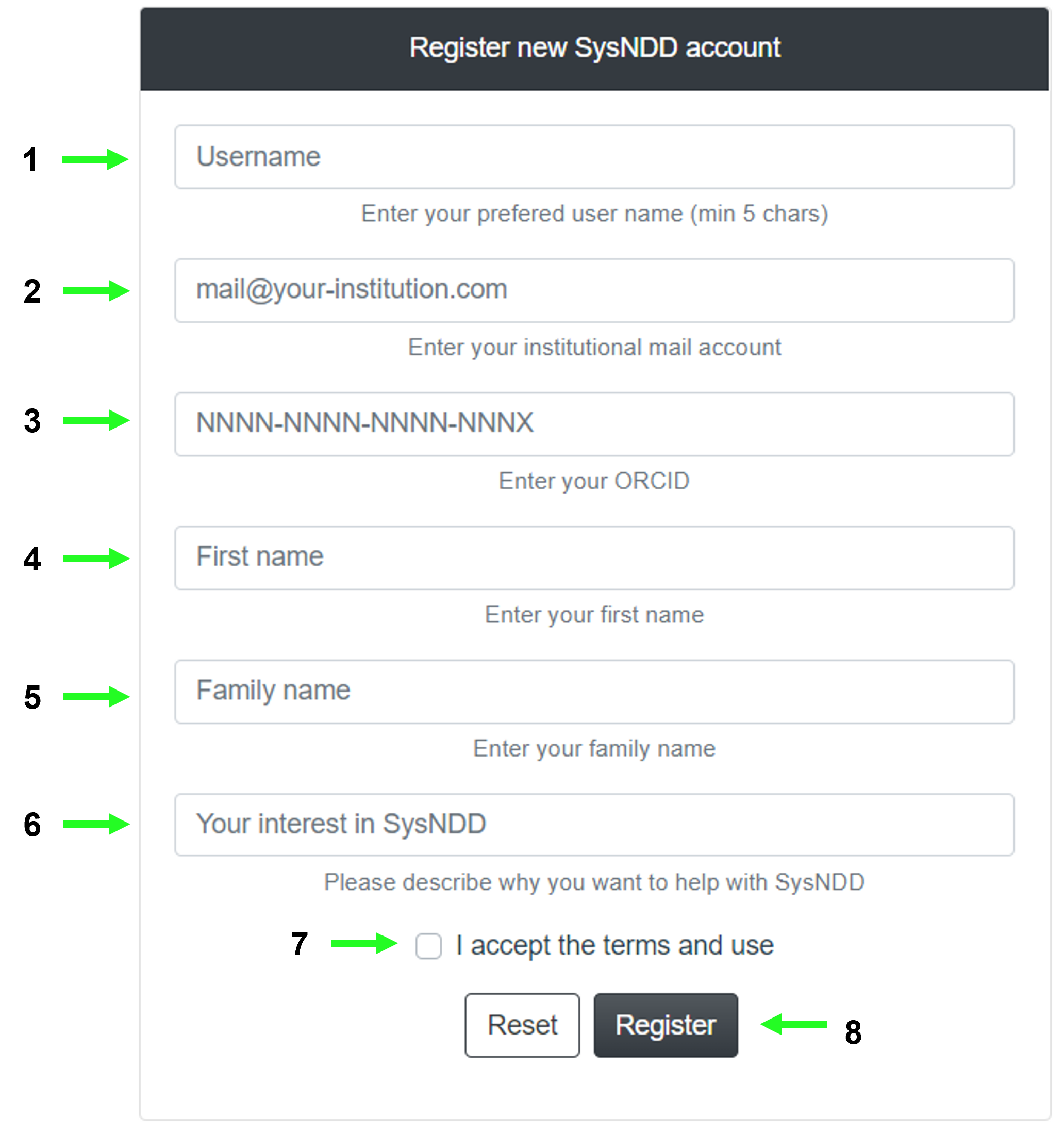
2.10.1 Register user page
This page can be used to apply for a SysNDD account by entering the following information:
- desired username
- institutional e-mail
- ORCID identifier
- first name
- family name
- description of your interest in SysNDD and why you want to participate in the curation effort
and (7) accepting the terms of use.
The (8) buttons allow resetting or submitting the form.
After submitting your application, the curators will receive an email to review it. You will receive an e-mail with your login information and instructions after your application has been confirmed.
2.11 Mobile website
The Vue.js framework enables native cross platform development. Together with the Bootstrap CSS library, this enables the SysNDD web app to integrate seamlessly on smaller mobile screens.
The layout breaks to mobile view at 768 pixels width and minimizes the navigation and footer menus:
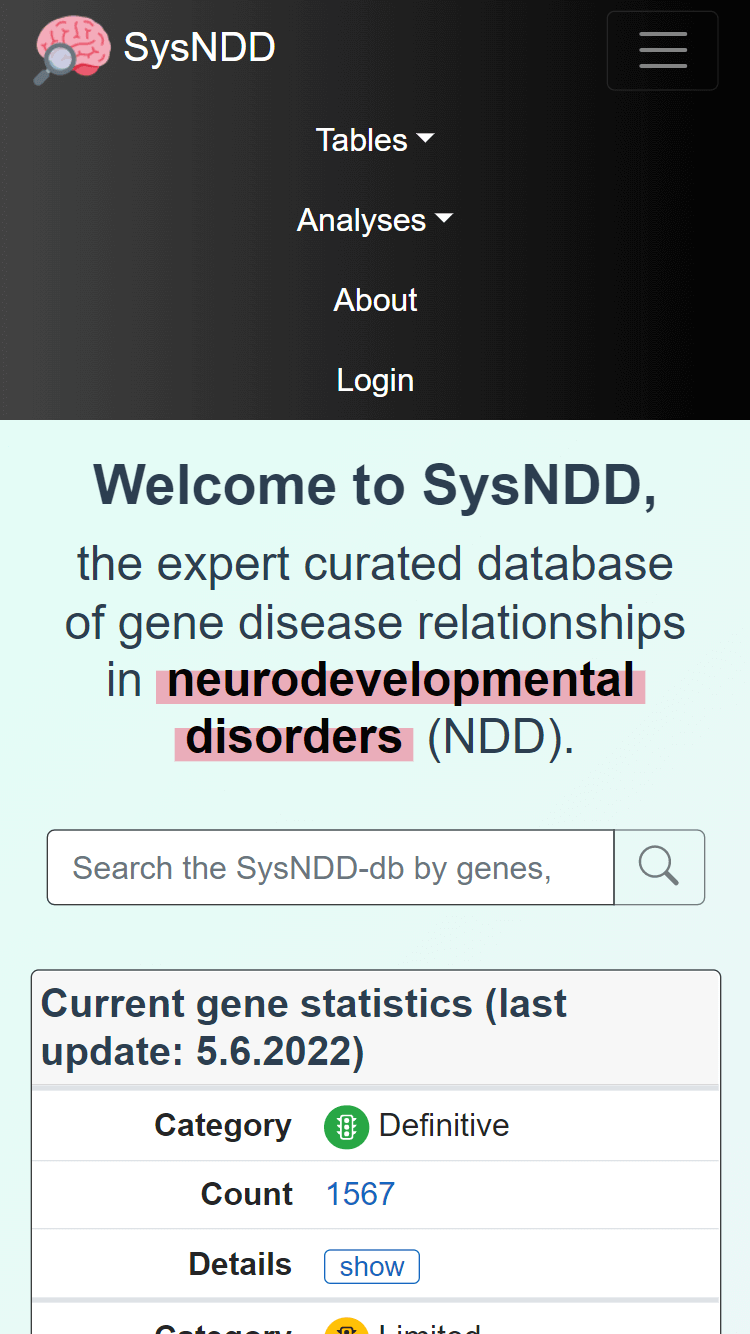
All tables in mobile views break to a stacked view (column names become the first column in a cell and values the second column) to best use display space:
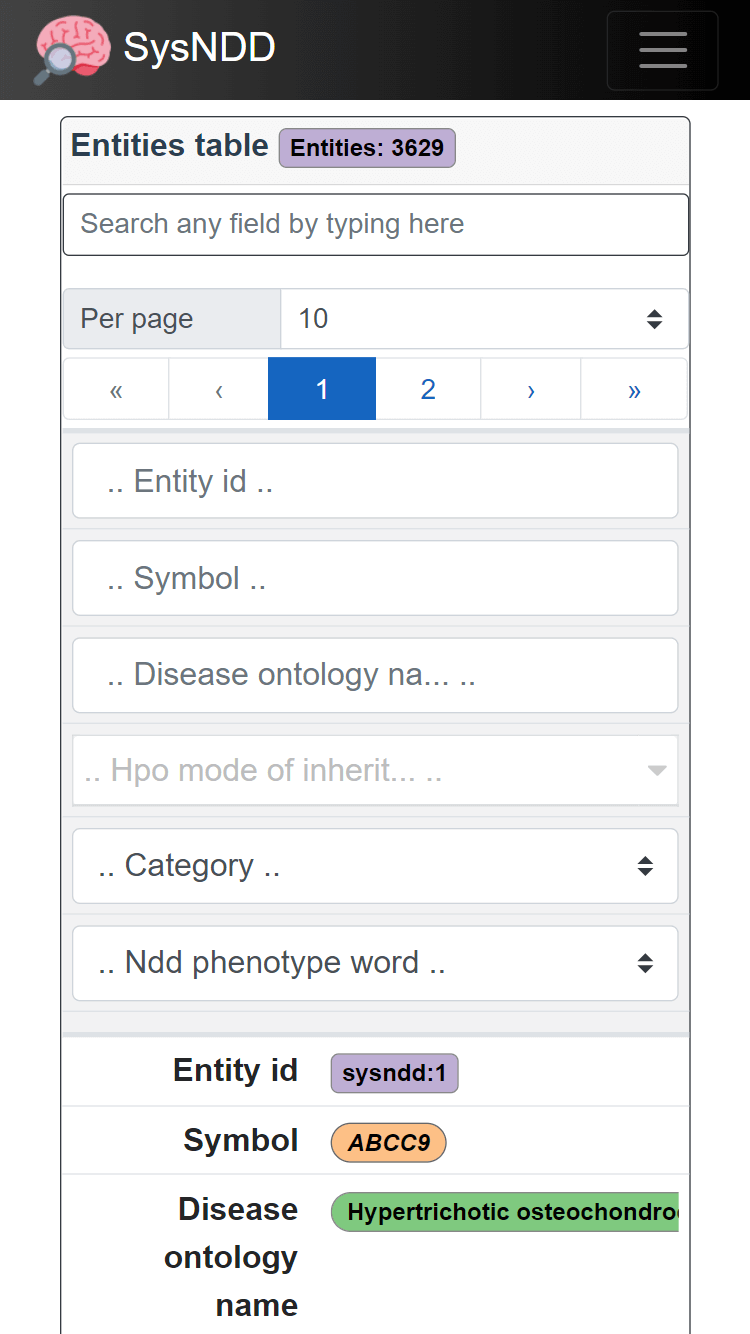
The table controls and search inputs are further displayed at the top in this view.
The Analyses pages on mobile are best viewed in landscape mode:
2.12 Progressive Web App (PWA)
The SysNDD web app can also be installed on mobile devices using the Progressive Web App (PWA) technology. This is supported in all Chromium-based modern browsers (Chrome, Edge, Opera, etc.) on all common operating systems (Windows, Linux, maxOS and Android). Additionally new Safari versions on iOS show some support for PWA.
PWAs are faster because they cache files. They offer more screen space for the app. Future integrations of this feature in SysNDD will enable offline use.
To install the PWA on Android devices follow these steps:
1) Visit the SysNDD website at https://sysndd.dbmr.unibe.ch/. You will see a message offering to add the PWA to your home screen:
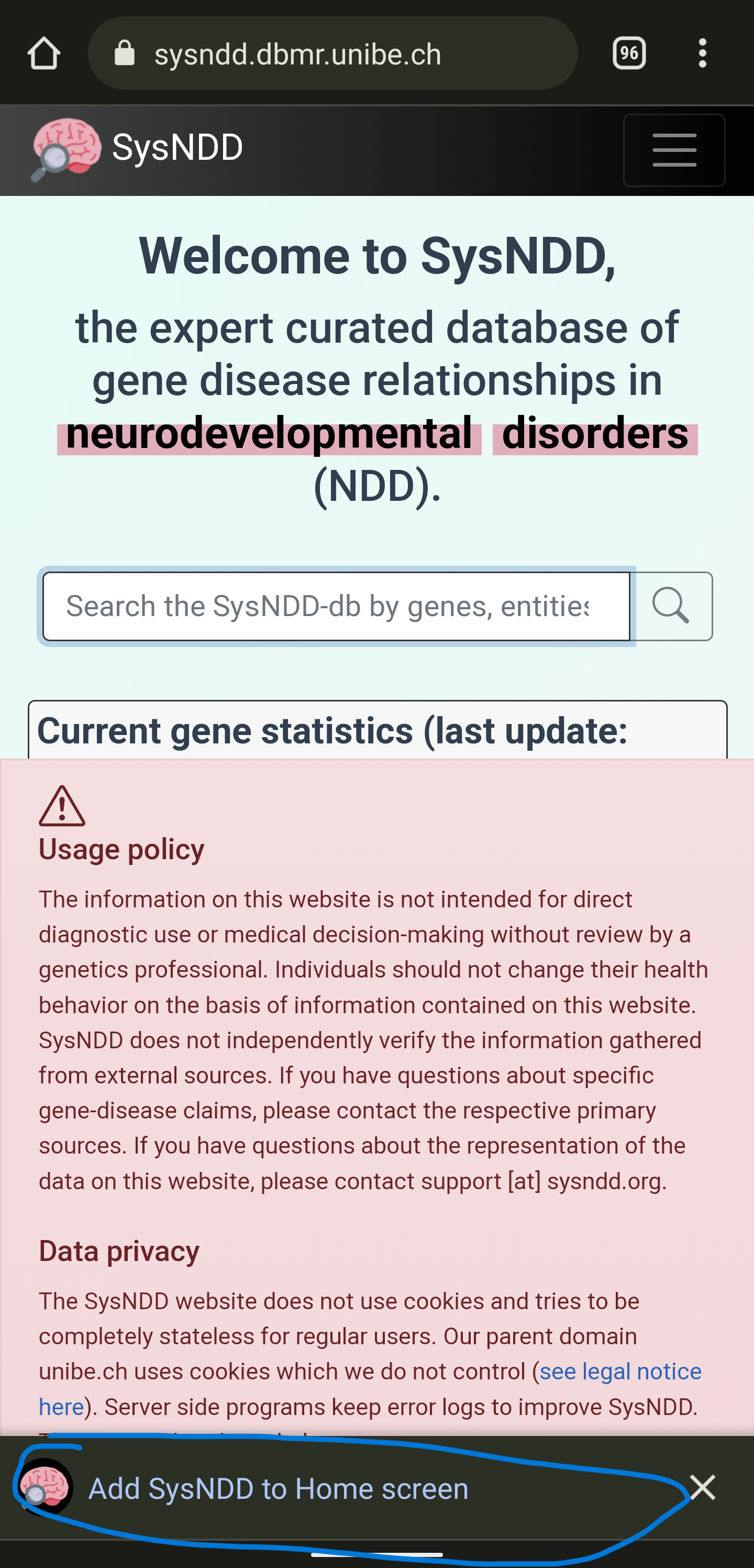
2) After clicking the previous message, confirm the installation by clicking “Install” in the following prompt:
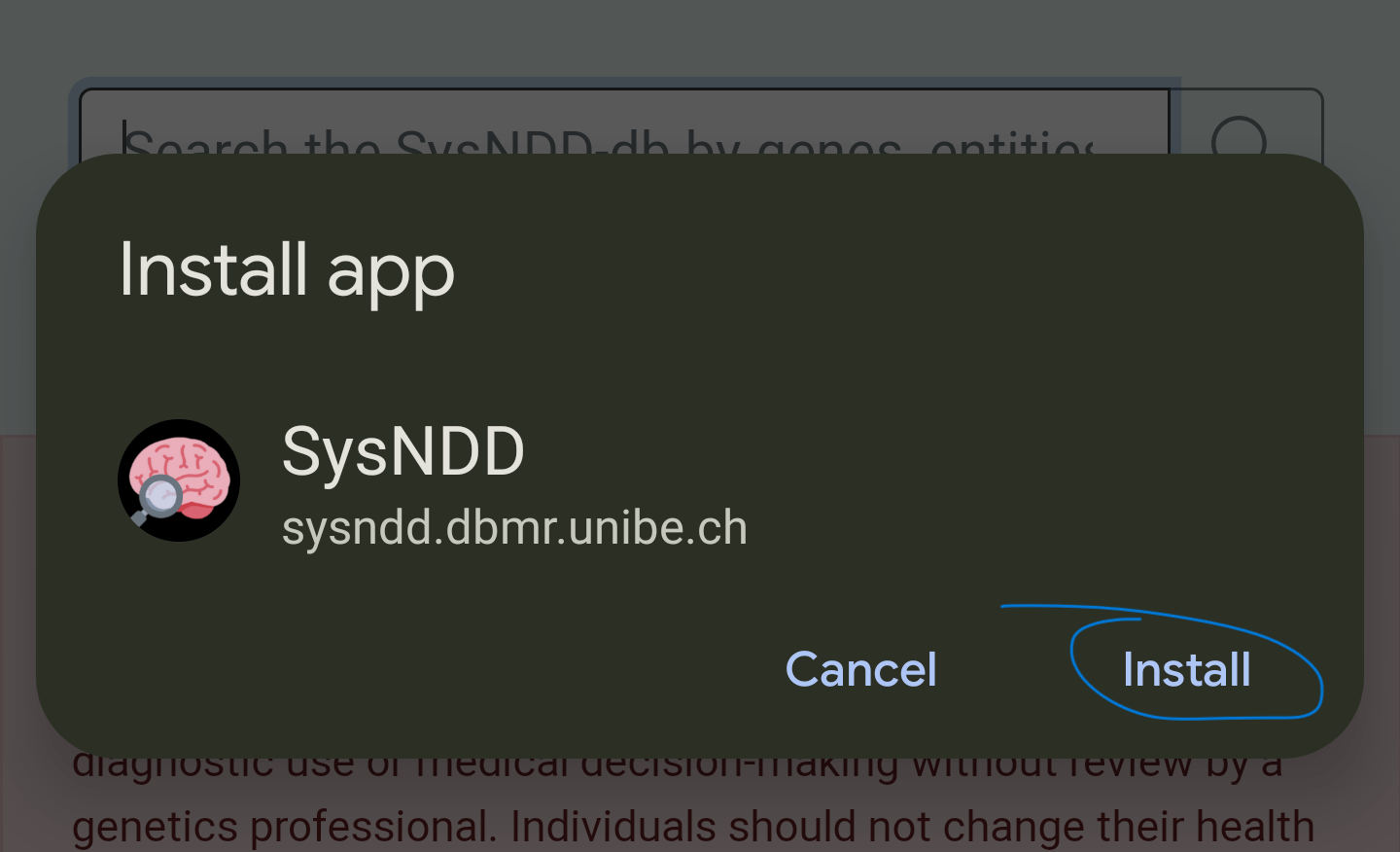
3) A message will confirm the installation:
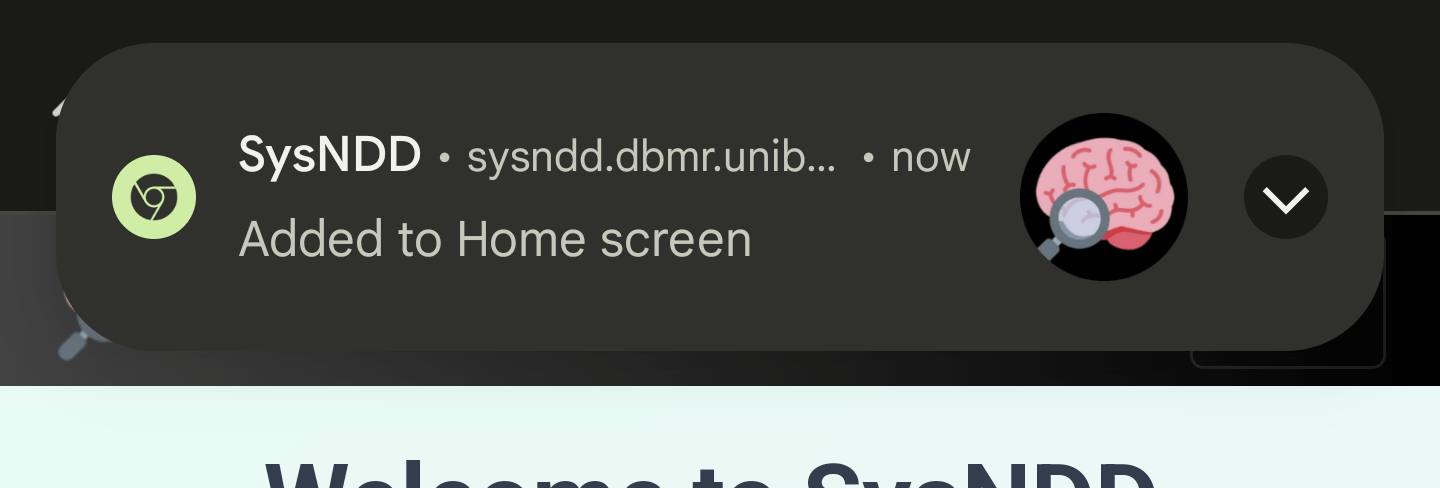
4) Following app symbol will be available on one of your screens:

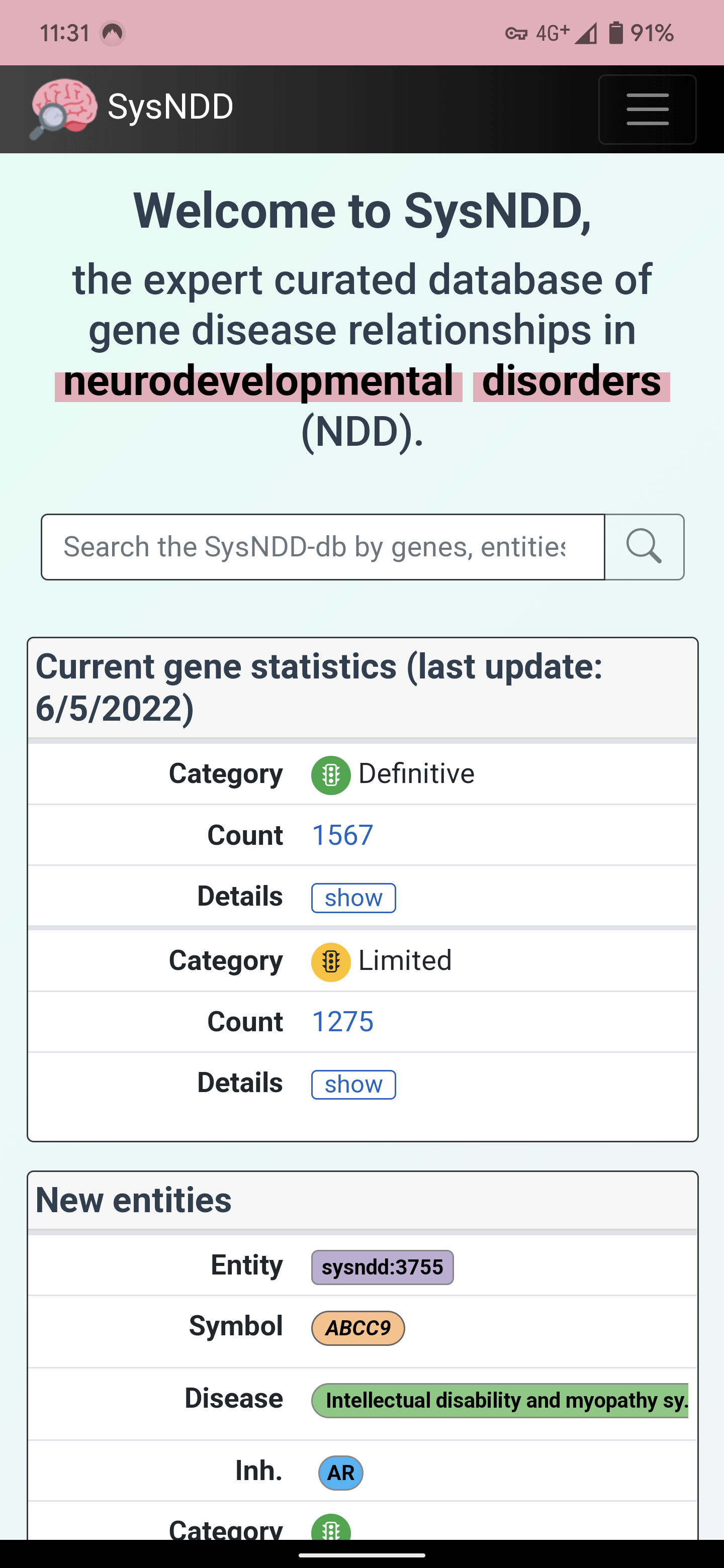
5) Clicking this will open SysNDD in PWA mode (no browser address bar, instead custom coloured top bar):
2.13 Performance
Modern Javascript frameworks like Vue.js, which we use for the SysNDD website, offer rich user experience. The generated single-page applications can be slower than server side rendered pages.
With SysNDD we are engaged to provide a fast user experience by reducing component and request sizes and applying techniques like lazy loading and code splitting in the frontend with parallelisation in the api.
A quick overview on the current website performance can be obtained on PageSpeed Insights (or “Lighthouse” in the chrome development console):
https://pagespeed.web.dev/report?url=https%3A%2F%2Fsysndd.dbmr.unibe.ch%2F
2.14 Security
SysNDD is engaged to offer highest security standards for all web tools. We use HTTPS with Transport Layer Security (TLS) and follow the Mozilla recommendations for web server settings.
A quick overview for our security settings for the SysNDD website can be obtained on Mozilla Observatory:
https://observatory.mozilla.org/analyze/sysndd.dbmr.unibe.ch